Training models with billions or trillions of parameters demands advanced parallel computing. Researchers must decide how to combine parallelism strategies, select the most efficient accelerated libraries, and integrate low-precision formats such as FP8 and FP4—all without sacrificing speed or memory.
There are accelerated frameworks that help, but adapting to these specific methodologies can significantly slow R&D, as users typically need to learn an entirely new codebase.
NVIDIA BioNeMo Recipes can simplify and accelerate this process by lowering the barrier to entry for large-scale model training. Using step-by-step guides built on familiar frameworks like PyTorch and Hugging Face (HF), we show how integrating accelerated libraries such as NVIDIA Transformer Engine (TE) unlocks speed and memory efficiency, scaling performance through techniques like Fully Sharded Data Parallel (FSDP) and Context Parallelism.
In this blog post, we demonstrate how to accelerate transformer-style AI models for biology by taking the Hugging Face ESM-2 protein language model with a native PyTorch training loop and:
- Accelerating it with TE. Integrating with FSDP2 for auto-parallelism. Showin sequence packing to achieve even greater performance.
All you need to get started is PyTorch, NVIDIA CUDA 12.8, and the following resources:
Integrating Transformer Engine into ESM-2
TE enables significant performance gains by optimizing transformer computations, particularly on NVIDIA GPUs. It can be integrated into existing training pipelines without requiring a complete overhaul of your datasets, data loaders, or trainers. This section shows how to incorporate TE into a model like ESM-2, drawing inspiration from the BioNeMo recipes.
In most use cases, using the ready-made TransformerLayer module from TE is straightforward. This encapsulates all fused TE operations and best practices into a single drop-in module, reducing boilerplate code and setup. The following snippet shows how we integrated TE in ESM-2. The full implementation can be found in the NVEsmEncoder class definition in bionemo-recipes.
import torchimport transformer_engine.pytorch as tefrom transformer_engine.common.recipe import Format, DelayedScalingclass MyEsmEncoder(torch.nn.Module): def __init__(self, num_layers, hidden_size, ffn_hidden_size, num_heads): super().__init__() self.layers = torch.nn.ModuleList([ te.TransformerLayer( hidden_size=hidden_size, ffn_hidden_size=ffn_hidden_size, num_attention_heads=num_heads, layer_type="encoder", self_attn_mask_type="padding", attn_input_format="bshd", # or 'thd', read below. window_size=(-1, -1), # disable windowed attention ) for _ in range(num_layers) ]) # Optionally add embedding, head, etc. def forward(self, x, attention_mask=None): for layer in self.layers: x = layer(x, attention_mask=attention_mask) return x# Layer configurationlayer_num = 8hidden_size = 4096sequence_length = 2048batch_size = 4ffn_hidden_size = 16384num_attention_heads = 32dtype = torch.bfloat16# Synthetic data (batch, seq, hidden) for bshd formatx = torch.rand(batch_size, sequence_length, hidden_size).cuda().to(dtype=dtype)attention_mask = torch.ones(batch_size, 1, 1, sequence_length, dtype=torch.bool).cuda()myEsm = MyEsmEncoder(layer_num, hidden_size, ffn_hidden_size, num_attention_heads)myEsm.to(dtype=dtype).cuda()fp8_recipe = DelayedScaling(fp8_format=Format.HYBRID)with te.fp8_autocast(enabled=True, fp8_recipe=fp8_recipe): y = myEsm(x, attention_mask=attention_mask)If your architecture deviates from a standard Transformer block, TE can still be integrated at the layer level. The core idea is to replace standard PyTorch modules (e.g., nn.Linear, nn.LayerNorm) with their TE counterparts and use FP8 autocasting to achieve maximum performance gains. TE provides several alternative implementations to common layers, such as Linear, fused LayerNormLinear, and attention modules like DotProductAttention and MultiheadAttention. For a complete list of supported modules, check the TE documentation.
Efficient sequence packing
Standard input data formats can be inefficient when samples have varying sequence lengths. For example, ESM-2 pretraining with a context length of 1,024 can consist of around 60% padding tokens, wasting compute on tokens that do not participate in the model’s attention mechanism. Internally, networks typically represent the hidden state of input sequences in a tensor with four dimensions: [batch size (B), max sequence length (S), number of attention heads (H), and head hidden dimension (D)], or BSHD.
As an alternative, modern attention kernels enable users to provide packed inputs without padding tokens, using index vectors to denote the boundaries between input sequences. Here, hidden states are represented by a flattened tensor of size [flattened input tokens (T), number of attention heads (H), head hidden dimension (D)], or THD. Figure 1 shows this format change, which results in less memory usage and faster token throughput by removing padding tokens (grey).
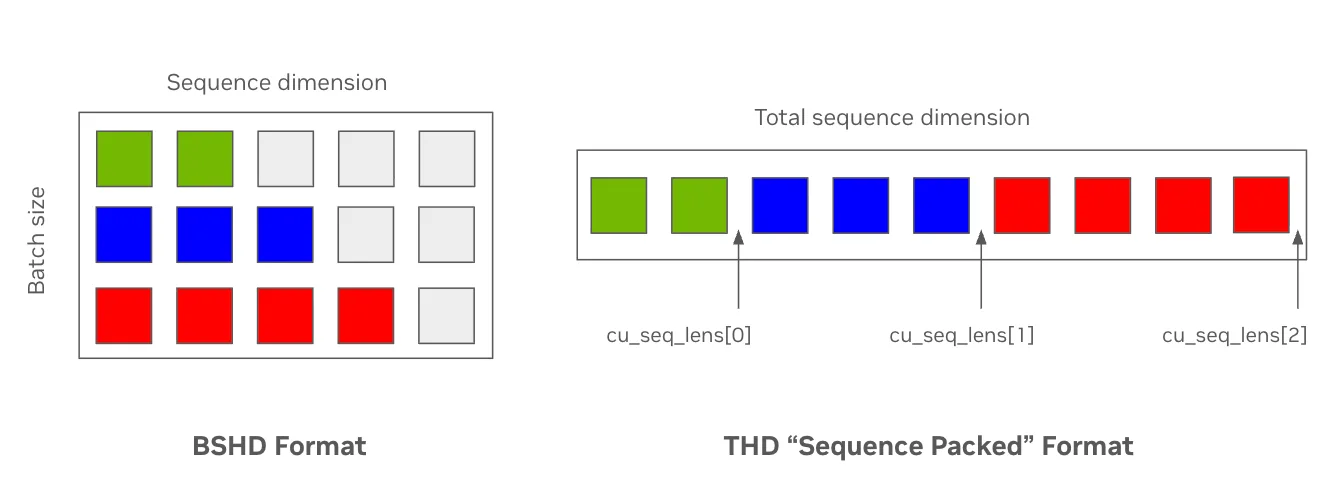
TE makes this optimization relatively simple by adding an attn_input_format parameter to relevant layers, which then accepts standard flash-attention-style cumulative sequence length keyword arguments (cu_seq_lens_q). These can be generated using THD-aware collators, such as Hugging Face’s DataCollatorWithFlattening, or the masking version implemented in BioNeMo Recipes.
def sequence_pack(input_ids, labels): # input_ids is a list of sequences: [(S1,), (S2,), ..., (SN,)] of shape (B,S) # Flatten and track sequence boundaries # Determine the length of each sequence sample_lengths = [len(sample) for sample in input_ids] # Flatten the input_ids and labels flat_input_ids = [token for sample in input_ids for token in sample] flat_labels = [label for sample in labels for label in sample] # Create a list of cumulative sums showing where the sequences start/stop # Note: for self attention cu_seqlens_q and cu_seqlens_kv will be the same cu_seqlens = torch.cumsum(torch.tensor([0] + sample_lengths), dim=0, dtype=torch.int32) max_length = max(sample_lengths) return { "input_ids": torch.tensor(flat_input_ids, dtype=torch.int64), "labels": torch.tensor(flat_labels, dtype=torch.int64), # These are the same kwargs used by `flash_attn_varlen_func`, etc. "cu_seqlens_q": cu_seqlens, "cu_seqlens_kv": cu_seqlens, "max_length_q": max_length, "max_length_kv": max_length, }TE and sequence packing on/off performance
Figure 2 shows the performance comparison, with a significant uplift in token throughput when TE is employed. This demonstrates TE’s ability to maximize the computational efficiency of your NVIDIA GPUs.
EvolutionaryScale integrated Transformer Engine across their next-generation models as well:
“ESM3 is the largest foundation model trained on biological data. Integrating the NVIDIA Transformer Engine was crucial to training it at this 98B parameter scale with high throughput and GPU utilization,” said Tom Sercu, co-founder and VP of Engineering at EvolutionaryScale. “The precision and speed of FP8 acceleration, combined with optimized kernels for fused layers, allow us to push the boundaries of compute and model scale across NVIDIA GPUs. This leads to emergent understanding of biology in our frontier models for the scientific community.”
Hugging Face interoperability
One of the key advantages of TE is its interoperability with existing machine learning ecosystems, including popular libraries like Hugging Face. This means you can use TE’s performance benefits even when working with models loaded from the Hugging Face Transformers library.
TE layers can be embedded directly inside a Hugging Face Transformers PreTrainedModel, and are fully compatible with AutoModel.from_pretrained. See the NVIDIA BioNeMo Collection on the Hugging Face Hub for pre-optimized models.
The process typically involves loading your Hugging Face model, then carefully identifying and replacing its standard PyTorch layers (such as nn.Linear, nn.LayerNorm, and nn.MultiheadAttention) with their TE-optimized counterparts. This often requires renaming some layers or a custom model wrapper to ensure the TE layers are correctly integrated into the model’s forward pass.
Get started
Our mission with BioNeMo Recipes is to make acceleration and scaling accessible for all foundation model builders. To help us build a more powerful and practical toolkit, we want to hear from you. We encourage you to try out the recipes and contribute by submitting a pull request or opening an issue on our GitHub.

